In this activity, the aim is to create a number of synthetic images as a practice for future testing of algorithms or simulations of optical phenomena in imaging systems.
Sample code written in Scilab that creates a circular aperture was given to us. The code produces the image:
This was performed by first creating two 2D arrays (X and Y) containing x and y coordinates ranging from -1 to 1 using the ndgrid() function. The number of x and y elements are set using the variables nx and ny respectively. With the line:
r = sqrt(X.^2 + Y.^2);
A matrix r is created that contains circle radius values. The value of r(x,y) is equal to the radius of a circle centered at (0,0) whose edge lies on (X(x,y),Y(x,y)). The matrix r represents the radius values of circles with varying sizes centered at the origin. When plotted, the matrix r looks like: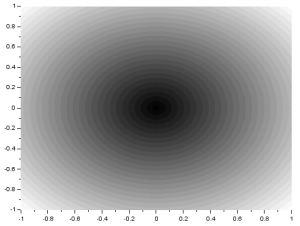
Then, a zero matrix A with the same size as r was created. With the line:
A (find(abs(r)<0.5) ) = 1;
Indices where r < 0.5 are chosen and the value of A at these indices is changed to 1. This will form a circle with a radius equal to the value in matrix r which is closest to but less than 0.5. For high nx and ny, this will produce a radius which is approximately 0.5.
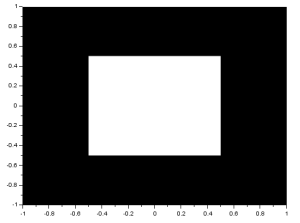
Other synthetic images can be made by varying A or r. To make a centered square aperture, I replaced last line above with:
A (find(abs(X)<0.5) ) = 1;
A (find(abs(Y)>0.5) ) = 0;
First, indices representing |x| < 0.5, the value of A is changed to 1. This will form a rectangle. The second line turns the image into a square by returning the values where |y| > 0.5 to 0. The resulting image is:
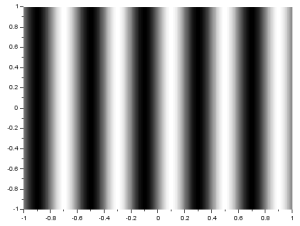
To produce a sinusoid along the x-direction or a corrugated roof, r was simply adjusted to:
r= sin(X*5*%pi);
and was plotted. The resulting image is:
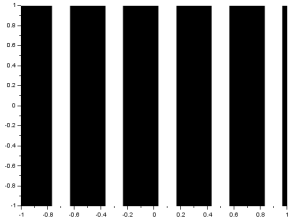
To produce grating along x, I simply added the line:
A (find(r>0.5) ) = 1;
to the code for the sinusoid. This is a discrete version of the sinusoid since certain values of r are changed to just either 1 or 0. The result is:
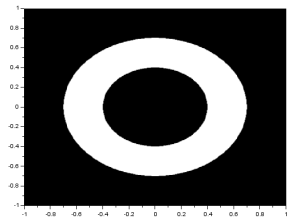
To produce an annulus, I simply adjusted the original code to produce two circles of different size. One circle made of 1 values and the other made of 0 values. The resulting image is:
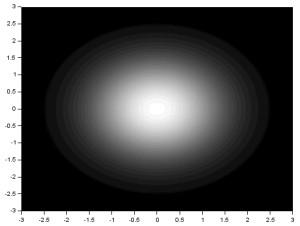
To create a circular aperture with Gaussian transparency, the range was changed to -3 to 3 and the matrix A in the original code was replaced with:
G = 1/sqrt(2*%pi)*exp(-(r.^2)/2);
G (find(r>2.5) ) = 0;
The first line is simply the equation for the Gaussian Distribution. The second line blackens the area outside of the circle with radius 0.8. The range was changed so that more shades of gray are used. The result is:
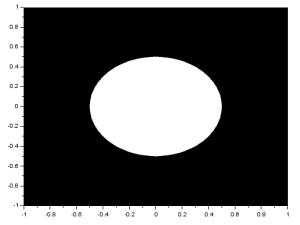
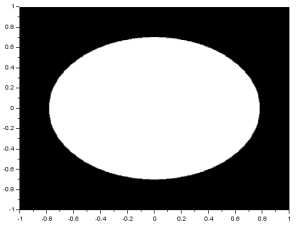
An ellipse can be made by simply multiplying either X or Y in the original code by a constant with a magnitude less than 1. The result is:
A cross can be made by adjusting the code for the square image. Replacing the lines assigning values to A with:
A (find(abs(X)<0.2) ) = 1;
A (find(abs(Y)<0.2) ) = 1;
forms the image:
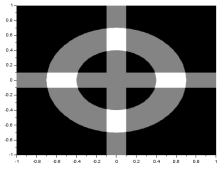
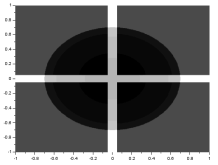
Using the matrix for the cross image and adding it to the annulus matrix, subtracting the gauss matrix from it, and multiplying it to the circle matrix, the following images were formed:
Figuring out how to produce each image was fun and maybe being new to programming in Scilab made it even more fun. Even figuring out some new functions like find() was enjoyable. Since I have produced all the required images and explored the outcomes of combining matrices for different images, i would rate myself with a grade of 10.
*ten tententenen ten tenenenenen ten ten ten teneeen
References:
M. Soriano, “A3 – Scilab Basics,” Applied Physics 186, National Institute of Physics, 2014.
Image References: