The aim in this activity is to segment images by picking a region of interest (ROI) and then processing the images based on features unique to the ROI.
For grayscale images where the ROI has a distinct grayscale range, segmentation can easily be done by using thresholding. For example, lets take the image:
Using Scilab, the grayscale histogram of the image read as I can be computed and plotted using the lines [1]:
[count, cells] = imhist(I, 256);
plot (cells, count);
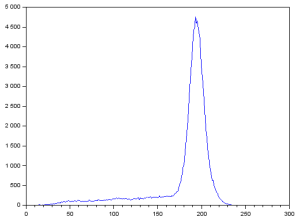
This results in:
The large peak corresponds to the background pixels which are lighter than the text. To segment the text, we can assume that using a threshold of 125 can be used since it is less than and far from the pixel values of the peak. This can be implemented using the lines [1]:
threshold = 125;
BW = I < threshold;
This results in:
where only the pixels having values less than the threshold of 125 are colored white. Using threshold values of 50, 100, 150, and 200, (left to right, up to down) I got:
We can see that using a lower threshold than needed will segment fewer details while using a higher threshold than needed will include too much unwanted details in the segmented image.
The problem of segmentation for colored images is more complicated. ROI’s that have unique features in a colored image might not be distinct after converting the image to grayscale [1]. Also, when segmenting images that have 3D objects, the variation in shading must be considered. Because of this, it is better to represent colors using the normalized chromaticity coordinates or NCC [1].
The NCC can be computed using the equation:
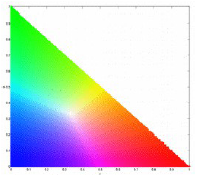
where . Since b is dependent on r and g, the chromaticity can be represented by using just the two coordinates r and g. I represents the brightness information [1]. The normalized chromaticity space is then shown in the plot:
where the x-axis is r and the y-axis is g [1].
A. Parametric Estimation
Segmentation can be done by calculating the probability that a pixel belongs to the color distribution of the ROI. To do this, we can crop a subregion of the ROI and compute its histogram. When normalized by the number of pixels, the histogram is already the probability distribution function (PDF) of the color. Assuming a Gaussian distribution for r and g, independently, of the cropped region, the means ,
, and standard deviations
,
can be computed [1]. The probability is then computed using the equation:
A similar equation can be used for p(g). The joint probability is then the product of p(r) and p(g) and can be plotted to produce a segmented image [1].
B. Non-Parametric Estimation
Histogram backprojection is a method where a pixel value in the image is given a value equal to its histogram value in the normalized chromaticity space. This can be used to segment images by manually looking up the histogram values. A 2D Histogram can be obtained by converting the r and g values to integers and binning the image values to a matrix. This can be implemented using the code [1]:
BINS = 180;
rint = round (r*(BINS-1) + 1);
gint = round (g*(BINS-1) + 1);
colors = gint(:) + (rint(:)-1)*BINS;
hist = zeros(BINS,BINS);
for row = 1:BINS
for col = 1:(BINS-row+1)
hist(row,col) = length( find(colors==( ((col + (row-1)*BINS)))));
end;
end;
where r and g are taken from the ROI. Then, the segmented image can be computed using the code [1]:
rint = round (r*(BINS-1) + 1);
gint = round (g*(BINS-1) + 1);
[x, y] = size(sub);
result = zeros(x, y);
for i = 1:x
for j = 1:y
result(i,j) = hist(rint(i,j), gint(i,j));
end;
end;
where r and g are taken from the subject image.
C. Results
The first image I segmented is a drawing of one of my favorite monsters from the game Monster Rancher, Mocchi:
souce: deviantart
The colors found on its head (green), lips (yellow), body (cream), and belly (cream) all appear solid. I cropped the following ROIs:
for the four parts. After applying parametric estimation, I got (left to right, up to down):
We can see that the corresponding body parts of Mocchi, depending on the cropped ROI, were successfully segmented. After looking closely at the image, I saw that there are actually distortions near the black pixels (Mocchi’s edges). These distortions are the reason why the segmented images do not exactly follow Mocchi’s shape.
Next, I segmented an image of pick-up sticks:
source: choicesdomatter.org
First, I used an ROI from a green stick that has minimal shading variations:
Using parametric estimation, I got the following segmented image:
We can see that some areas are not segmented perfectly. These areas are those of really bright or really dark shading. Using an ROI from a stick with more shading variations:
I got:
We can see that the green sticks are clearly identified by the white pixels, however, other areas are no longer black. This is because the features of the new ROI are no longer unique to just the green sticks. For the other colors, I will continue with using ROIs taken from sticks with minimal shading variations:
I will also apply non-parametric estimation. First, I plotted the histogram of the ROIs, respectively:
If we look at the plot of the normalized chromaticity space again, we can see that the location of the peaks are located inside the corresponding expected regions. Therefore we can say that the histograms are correct. For the green, red, yellow, and blue ROIs, respectively, I got a segmented image using parametric estimation (left) and non-parametric (right) estimation:
green:
red:
yellow:
blue:
We can see that using parametric estimation segments the corresponding pick-up sticks more accurately than using non-parametric estimation. For this case, using non-parametric estimation identifies fewer pixels to be similar to the ROI since it directly uses the histogram instead of using a probability distribution. Therefore it is not suitable for use when segmenting images with objects that have really small widths such as these pick-up sticks. After using non-parametric estimation on the image of Mocchi using the same ROIs shown in the first part of the results, I got:
We can see that the segmentation is still successful since the parts corresponding to the different ROIs are large enough.
This activity was fun because I can instantly be gratified by the results since they can be confirmed by just looking at the segmented images.

Since I was able to obtain all the required images, I will rate myself with 10/10. Also, thanks to John Kevin Sanchez, Ralph Aguinaldo, and Gio Jubilo for helping me in correcting my equations and understanding backprojection.
References:
- M. Soriano, “A7 – Image Segmentation,” Applied Physics 186, National Institute of Physics, 2014.
Image Reference: